Many of the most devastating human diseases are heritable. Whether an individual develops schizophrenia, obesity, diabetes, or autism depends more on their genes (and epi-genome) than it does on any other factor. However, current genetic models do not explain all of this heritability. Twin studies show that schizophrenia, for example, is 80% heritable (over a broad cohort of Americans), but our best genetic model only explains ~9% of variance in cases. To be fair, we selected a dramatic example here (models for other diseases perform far better). Still, the gap between heritability and prediction stands in the way of personalized genetic medicine.
We are launching Tabula Bio to close this gap. We have a three-part thesis on how to approach this.
- The path forward is machine learning. The human genome is staggeringly complex. In the 20 years since the Human Gnome Project, much progress has been made, but we are still entirely short of a mechanistic, bottom-up model that would allow anything like disease prediction. Instead, we have to rely on statistical modeling. And statistical methods are winning over expert systems across domains. Expert-system chess AIs have fallen to less-opinionated ML, syntax-aware NLP models were left in the dust by LLMs, and more recently constraint-based robotics is being replaced by pixel-to-control machine learning. We are betting on the same trend extending to biology.
- The core problem is limited data and large genomes. The human genome contains 3 billion base pairs, while labeled biobank datasets (genomes and disease diagnoses) are numbered in the hundreds of thousands. Complex models thus hopelessly overfit and fail to generalize. Additionally, the human genome is highly repetitive and much of it likely has no relation to phenotype. Because of these problems, we can’t simply train high-parameter black-box models (or treat DNA like language in a language model).
- Given 1 and 2, novel ML architectures will be required. This is consistent with other breakthroughs in AI. Different problems require different inductive biases. An ML architecture for disease prediction should:
- Make use of unlabeled genomics data (human and non-human) as well as homogeneous biobank data. Much genetic coding is conserved across species. Ignoring this is leaving data on the table.
- Include priors from human expert research. It would be wonderful to start from a blank slate (we named the company Tabula for a reason). But, for example, we know that DNA is a 3D molecule and that the distance (in 3D space) between genes and regulatory sequences matters. An architecture needs to start from this premise.
- Include epigenetic data. We know it’s part of the story (maybe a large part).
We’re interested in probabilistic programming as a method to build such a model.
This is an ambitious idea, and we’re just getting started. But if we look into the future, to a world where humans have closed the heritability gap and personalized genetic medicine has eradicated great swaths of disease, it’s hard to imagine we did not get there via an effort like this.
Our team is currently Ammon Bartram, who was previously cofounder of Triplebyte and Socialcam, and Michael Poon, who has spent the past several years working on polygenic screening, an early employee at Twitch, and studied CS at MIT. We’re honored to have Michael Seibel and Emmett Shear among our initial investors.
Please reach out to us at team@tabulabio.com to help us harden our hypotheses or collaborate together on this mission! We're especially interested in talking with ML engineers interested in genomics and bioinformaticians interested in ML.
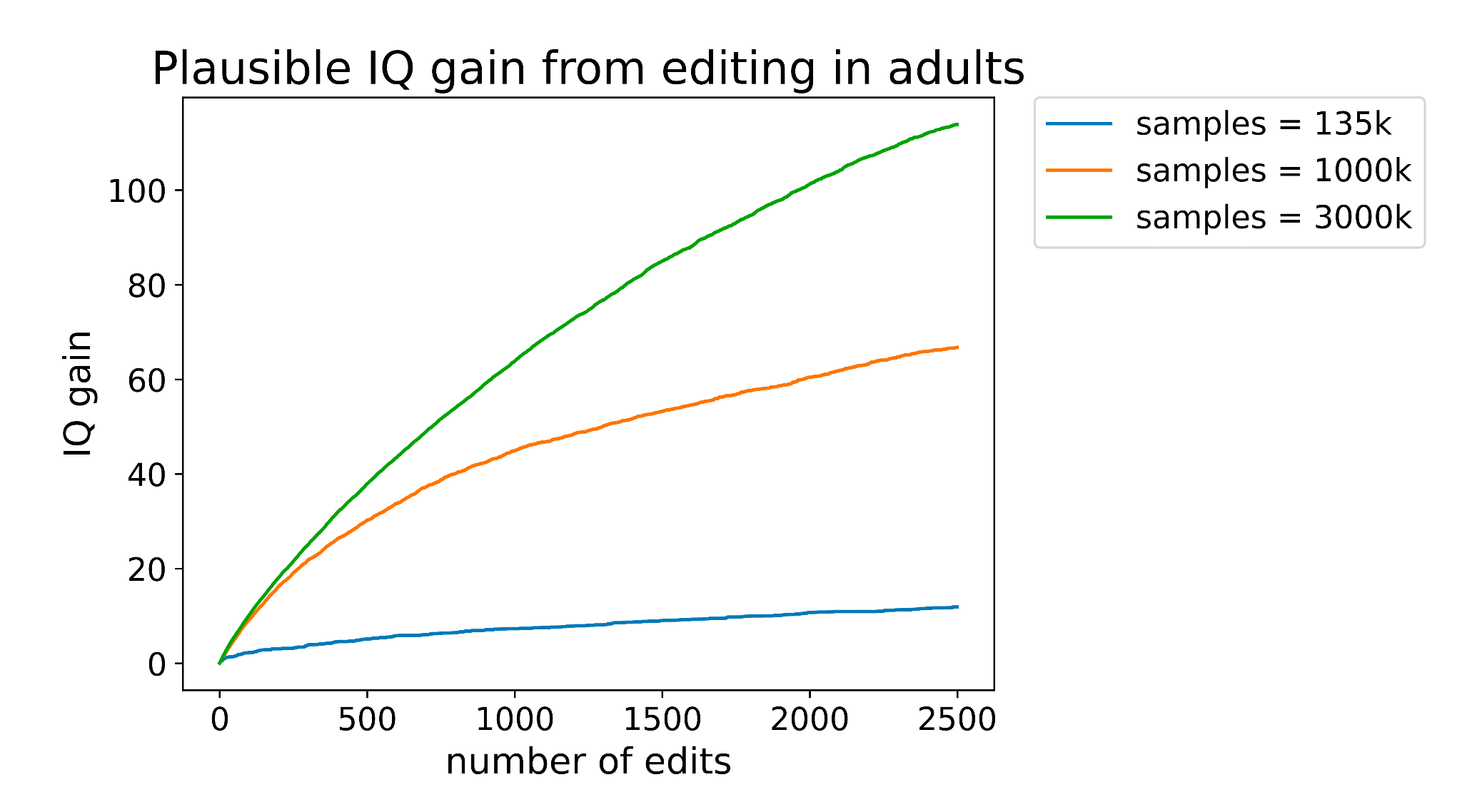

Why should we expect novel ML architectures to perform better than current methods like LASSO regression? If your task is disease risk prediction, this falls under "variance within the human distribution" which, as far as I know, should be explained mostly by additive effects of SNPs. Current missing heritability is more of an issue of insufficient data (like you say) and rare variants rather than an inefficiency of our models. Steve Hsu had a paper about why we should actually theoretically expect LASSO regression to be efficient here.
My impression is generally that genomics is one of the few areas in bio where fancy ML is ill-suited for the problem (specifically polygenic trait prediction). I'm curious if there's some premise I have that Tabula disagrees with that makes this venture promising.
ML arguments can take more data as input. In particular, the genomic sequence is not a predictor used in LASSO regression models: the variants are just arbitrarily coded as 0,1, or 2 alternative allele count. The LASSO models have limited ability to pool information across variants or across data modes. ML models like this one can (in theory) predict effects of variants just based off their sequence on data like RNA-sequencing (which shows which genes are actively being transcribed). That information is effectively pooled across variants and ties genomic s... (read more)