I'm a software developer by training with an interest in genetics. I currently run a startup working on multiplex gene editing technology.
Posts
Wikitag Contributions
I'm being gaslit so hard right now
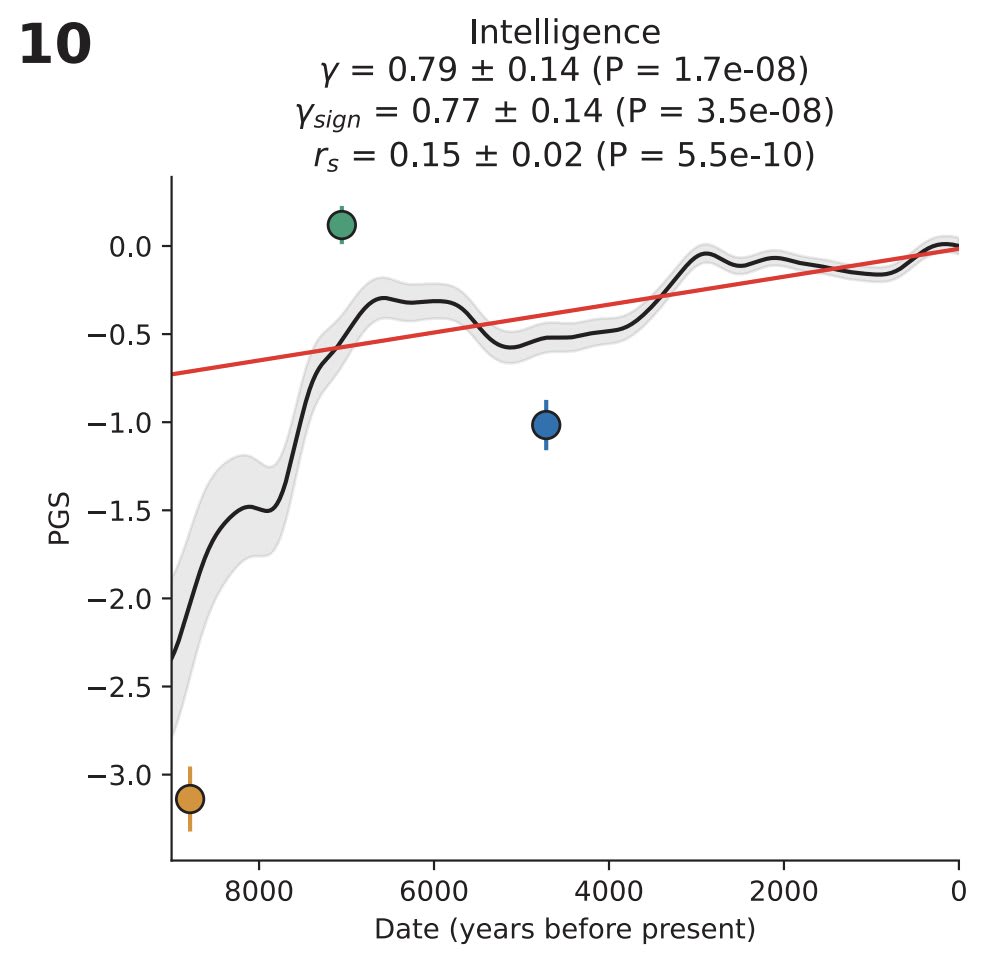
One data point that's highly relevant to this conversation is that, at least in Europe, intelligence has undergone quite significant selection in just the last 9000 years. As measured in a modern environment, average IQ went from ~70 to ~100 over that time period (the Y axis here is standard deviations on a polygenic score for IQ)
The above graph is from David Reich's paper
I don't have time to read the book "Innate", so please let me know if there are compelling arguments I am missing, but based on what I know the "IQ-increasing variants have been exhausted" hypothesis seems pretty unlikely to be true.
There's well over a thousand IQ points worth of variants in the human gene pool, which is not what you would expect to see if nature had exhaustively selected for all IQ increasing variants.
Unlike traits that haven't been heavily optimized (like resistance to modern diseases)
Wait, resistance to modern diseases is actually the single most heavily selected for thing in the last ten thousand years. There is very strong evidence of recent selection for immune system function in humans, particularly in the period following domestication of animals.
Like there has been so much selection for human immune function that you literally see higher read errors in genetic sequencing readouts in regions like the major histocompatibility complex (there's literally that much diversity!)
but suggests the challenge may be greater than statistical models indicate, and might require understanding developmental pathways at a deeper level than just identifying associated variants.
If I have one takeaway from the last ten years of deep learning, it's that you don't have to have a mechanistic understanding of how your model is solving a problem to be able to improve performance. This notion that you need a deep mechanical understanding of how genetic circuits operate or something is just not true.
What you actually need to do genetic engineering is a giant dataset and a means of editing.
Statistical methods like finemapping and adjusting for population level linkage disequilibrium help, but they're just making your gene editing more efficient by doing a better job of identifying causal variants. They don't take it from "not working" to "working".
Also if we look at things like horizontal gene transfer & shifting balance theory we can see these as general ways to discover hidden genetic variants in optimisation and this just feels highly non-trivial to me? Like competing against evolution for optimal information encoding just seems really difficult apriori? (Not a geneticist so I might be completely wrong here!)
Horizontal gene transfer doesn't happen in humans. That's mostly something bacteria do.
There IS weird stuff in humans like viral DNA getting incorporated into the genome, (I've seen estimates that about 10% of the human genome is composed of this stuff!) but this isn't particularly common and the viruses often accrue mutations over time that prevents them from activating or doing anything besides just acting like junk DNA.
Occasionally these viral genes become useful and get selected on (I think the most famous example of this is some ancient viral genes that play a role in placental development), but this is just a weird quirk of our history. It's not like we're prevented from figuring out the role of these genes in future outcomes just because they came from bacteria.
Sorry, I've been meaning to make an update on this for weeks now. We're going to open source all the code we used to generate these graphs and do a full write-up of our methodology.
Kman can comment on some of the more intricate details of our methodology (he's the one responsible for the graphs), but for now I'll just say that there are aspects of direct vs indirect effects that we still don't understand as well as we would like. In particular there are a few papers showing a negative correlation between direct and indirect effects in a way that is distinct for intellligence (i.e. you don't see the same kind of negative correlation for educational attainment or height or anything like that). It's not clear to us at this exact moment what's actually causing those effects and why different papers disagree on the size of their impact.
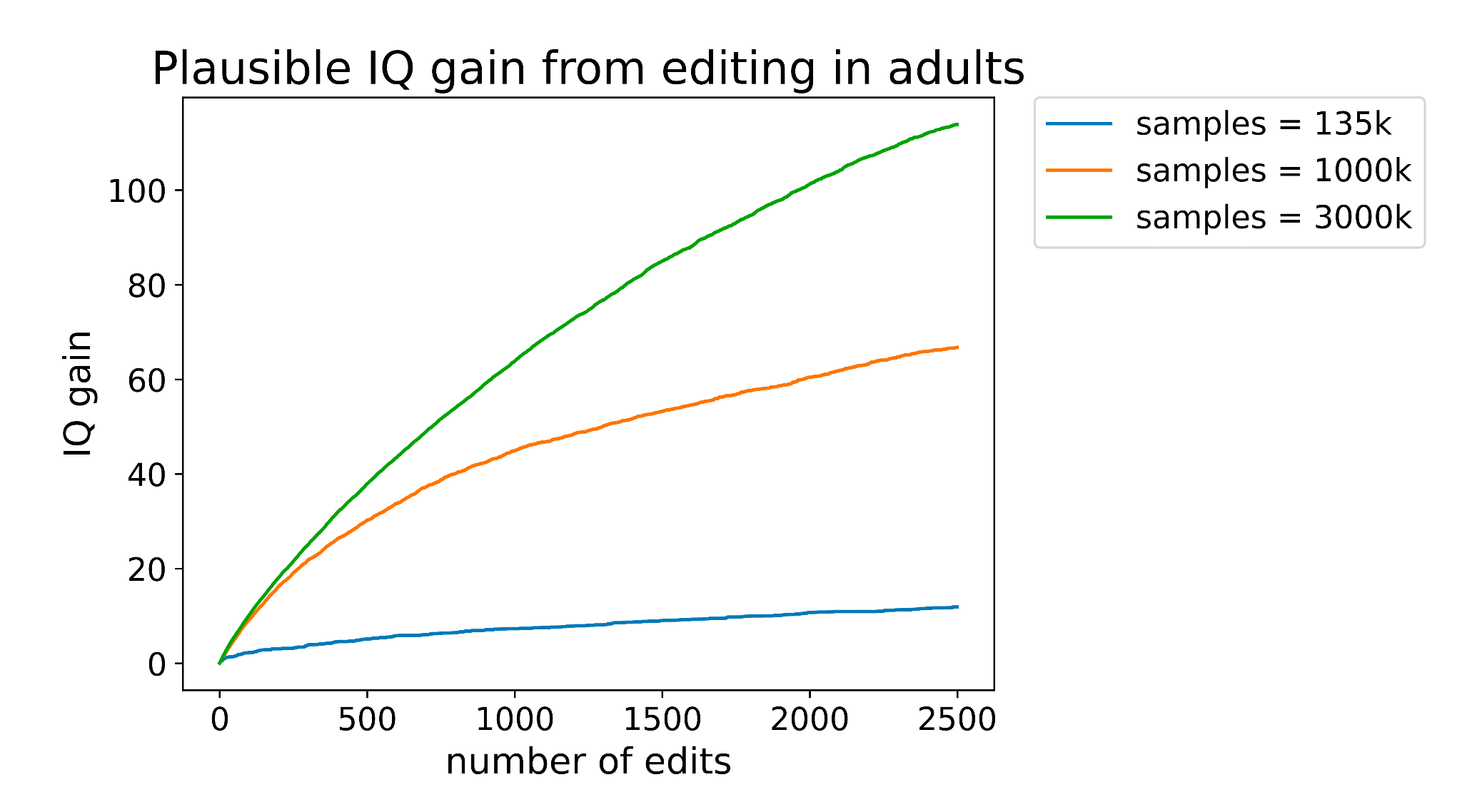
In the latest versions of the IQ gain graph we've made three updates:
- We fixed a bug where we squared a term that should not have been squared (this resulted in a slight reduction in the effect size estimate)
- We now assume only ~82% of the effect alleles are direct, further reducing benefit. Our original estimate was based on a finding that the direct effects of IQ account for ~100% of the variance using the LDSC method. Based on the result of the Lee et al Educational Attainment 4 study, I think this was too optimistic.
- We now assume our predictor can explain more of the variance. This update was made after talking with one of the embryo selection companies and finding their predictor is much better than the publicly available predictor we were using
The net result is actually a noticeable increase in efficacy of editing for IQ. I think the gain went from ~50 to ~85 assuming 500 edits.
It's a little frustrating to find that we made the two mistakes we did. But oh well; part of the reason to make stuff like this public is so others can point out mistakes in our modeling. I think in hindsight we should have done the traditional academic thing and ran the model by a few statistical geneticists before publishing. We only talked to one, and he didn't get into enough depth for us to discover the issues we later discovered.
I've been talking to people about this today. I've heard from two separate sources that it's not actually buyable right now, though I haven't yet gotten a straight answer as to why not.
Congrats on the new company! I think this is potentially quite an important effort, so I hope anyone browing this forum who has a way to get access to biobank data from various sources will reach out.
One of my greatest hopes for these new models is that they will provide a way for us to predict the effects of novel genetic variants on traits like intelligence or disease risk.
When I wrote my post on adult intelligence enhancement at the end of 2023, one of the biggest issues was just how many edits we would need to make to achieve significant change in intelligence.
Neither delivery nor editing tech is good enough yet to make anywhere close to the number of edits needed for adult cognitive enhancement. But it's plausible that there exist a set of gene edits such that just 10 or 20 changes would have a significant impact.
One of my greatest hopes is that these foundation models will generalize well enough out-of-distribution that we can make reasonably accurate predictions about the effect of NEW genetic variants on traits like intelligence of health. If we can, (and ESPECIALLY if we can also predict tolerance to these edits), it could 10x the impact of gene editing.
This may end up being one of the most important technologies for adult cognitive enhancement. So I hope anyone who might be able to help with data access reaches out to you!
I’ve checked. Have heard from multiple people they “it’s not for sale in reality”
I don’t have any details yet. But obviously am interested.
I certainly hope we can do this one day. The biobanks that gather data used to make the predictors we used to identify variants for editing don't really focus on much besides disease. As a result, our predictors for personality and interpersonal behavior aren't yet very good.
I think as the popularity of embryo selection continues to increase, this kind of data will be gathered in exponentially increasing volumes, at which point we could start to think about editing or selecting for the kinds of traits you're describing.
There will be an additional question to what degree parents will decide to edit for those traits. We're going to have a limited budget for editing and for selection for quite some time, so parents will have to choose to make their child kinder and more benificent to others at the expense of some other traits. The polygenicity of those personality traits and the effect sizes of the common alleles could have a very strong effect on parental choices; if you're only giving up a tiny bit to make your child kinder then I think most parents will go for it. If it's a big sacrifice because it requires like 100 edits, I think far fewer will do so.
It may be that benificence towards others will make these kinds of children easier to raise as well, which I think many parents would be interested in.
In the last year it has really hit me at a personal level what graphs like these mean. I'm imagining driving down to Mountain View and a town once filled with people who had "made it" and seeing a ghost town. No more jobs, no more prestige, no more promise of a stable life. As the returns to capital grow exponentially and the returns to labor decline to zero, the gap between the haves and the have-nots will only grow.
If someone can actually get superintelligence to do what they want, then perhaps universal basic income can at the very least prevent actual starvation and maybe even provide a life of abundance.
But I can't help but feeling such a situation is fundamentally unstable. If the government's desires become disconnected from those of the people at any point, by what mechanism can balance be restored?
In the past the government was fundamentally reliant on its citizens for one simple reason; citizens produced taxable revenue.
That will no longer be the case. Every country will become a petro state on steroids.
I spoke with one of the inventors of bridge recombinases at a dinner a few months ago and (at least according to him), they work in human cells.
I haven't verified this independently in my lab, but it's at least one data point.
On a broader note, I find the whole field of gene therapy very confusing. In many cases it seems like there are exceptionally powerful tools that are being ignored in favor of sloppy, dangerous, imprecise alternatives.
Why are we still using lentiviral vectors to insert working copies of genes when we can usually just fix the broken gene using prime editors?
You look at gene therapies like Casgevy for sickle cell and they just make no fucking sense.
Sickle cell is predominantly cause by an adenine to thymine swap at the sixth codon in the HBB gene. Literally one letter change at a very well known spot in one protein.
You'd think this would be a perfect use case for gene editing, right? Just swap out that letter and call it a day!
But no. This is not how Casgevy works. Instead, Casgevy works by essentially flipping a switch to make the body stop producing adult hemoglobin and start producing fetal hemoglobin.
Fetal hemoglobin doesn't sickle, so this fixes sickle cell. But like... why? Why not just change the letter that's causing all the problems in the first place?
It's because they're using old school Cas9. And old school Cas9 editing is primarily used to break things by chopping them in half and relying on sloppy cellular repair processes like non-homologous end joining to stitch the DNA back together in a half-assed way that breaks whatever protein is being produced.
And that's exactly what Casgevy does; it uses Cas9 to induce a double stranded break in BCL11A, a zinc finger transcription factor that normally makes the cells produce adult hemoglobin instead of the fetal version. Once BCL11A is broken, the cells start producing fetal hemoglobin again.
But again...
Why?
Prime editors are very good at targeting the base pair swap needed to fix sickle cell. They've been around for SIX YEARS. They havery extremely low rates of off-target editing. Their editing efficiency is on-par with that of old-school Cas9. And they have lower rates of insertion and deletion errors near the edit site. So why don't we just FIX the broken base pair instead of this goofy work-around?
Yet the only thing I can find online about using them for sickle cell is a single line announcement from Beam Therapeutics that vaguely referecing a partnership with prime medicine that MIGHT use them for sickle cell.
This isn't an isolated incident either. You go to conferences on gene editing and literally 80% of academic research is still using sloppy double strand breaking Cas9 to do editing. It's like if all the electric car manufacturers decided to use lead acid batteries instead of lithium ion.
It's just too slow. Everything is too fucking slow. It takes almost a decade to get something from proof of concept to commercial product.
This, more than anything, is why I hope special economic zones like Prospera win. You can take a therapy from animal demonstration to commercial product in less than a year for $500k-$1 mil. If we had something like that in the US there would be literally 10-100x more therapeutics available.
So in theory I think we could probably validate IQ scores of up to 150-170 at most. I had a conversation with the guys from Riot IQ and they think that with larger sample sizes the tests can probably extrapolate out that far.
We do have at least one example of a guy with a height +7 standard deviations above the mean actually showing up as a really extreme outlier due to additive genetic effects.
The outlier here is Shawn Bradley, a former NBA player. Study here
Granted, Shawn Bradley was chosen for this study because he is a very tall person who does not suffer from pituitary gland dysfunction that affects many of the tallest players. But that's actually more analogous to what we're trying to do with gene editing; increasing additive genetic variance to get outlier predispositions.
I agree this is not enough evidence. I think there are some clever ways we can check how far additivity continues to hold outside of the normal distribution, such as checking the accuracy of predictors at different PGSes, and maybe some clever stuff in livestock.
This is on our to-do list. We just haven't had quite enough time to do it yet.
There are some, but not THAT many. Estimates from EA4, the largest study on educational attainment to date, estimated the indirect effects for IQ at (I believe) about 18%. We accounted for that in the second version of the model.
It's possible that's wrong. There is a frustratingly wide range of estimates for the indirect effect sizes for IQ in the literature. @kman can talk more about this, but I believe some of the studies showing larger indirect effects get such large numbers because they fail to account for the low test-retest reliability of the UK biobank fluid intelligence test.
I think 0.18 is a reasonable estimate for the proportion of intelligence caused by indirect effects. But I'm open to evidence that our estimate is wrong.