I'm a bit skeptical of AlphaFold 3
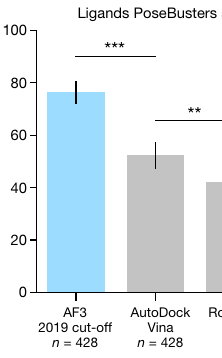
(also on https://olegtrott.substack.com) So this happened: DeepMind (with 48 authors, including a new member of the British nobility) decided to compete with me. Or rather, with some of my work from 10+ years ago. Apparently, AlphaFold 3 can now predict how a given drug-like molecule will bind to its target protein. And it does so better than AutoDock Vina (the most cited molecular docking program, which I built at Scripps Research): On top of this, it doesn’t even need a 3D structure of the target. It predicts it too! But I’m a bit skeptical. I’ll try to explain why. Consider a hypothetical scientific dataset where all data is duplicated: Perhaps the scientists had trust issues and tried to check each others’ work. Suppose you split this data randomly into training and test subsets at a ratio of 3-to-1, as is often done: Now, if all your “learning” algorithm does is memorize the training data, it will be very easy for it to do well on 75% of the test data, because 75% of the test data will have copies in the training data. Scientists mistrusting each other are only one source of data redundancy, by the way. Different proteins can also be related to each other. Even when the sequence similarity between two proteins is low, because of evolutionary pressures, this similarity tends to be concentrated where it matters, which is the binding site. Lastly, scientists typically don’t just take random proteins and random drug-like molecules, and try to determine their combined structures. Oftentimes, they take baby steps, choosing to study drug-like molecules similar to the ones already discovered for the same or related targets. So there can be lots of redundancy and near-redundancy in the public 3D data of drug-like molecules and proteins bound together. Long ago, when I was a PhD student at Columbia, I trained a neural network to predict protein flexibility. The dataset I had was tiny, but it had interrelated proteins already: With a larger dataset, du
Patents are valid for about 20 years. But Bengio et al used NNs to predict the next word back in 2000:
https://papers.nips.cc/paper_files/paper/2000/file/728f206c2a01bf572b5940d7d9a8fa4c-Paper.pdf
So this idea is old. Only some specific architectural aspects are new.