This post is based on chapter 15 of Uri Alon’s book An Introduction to Systems Biology: Design Principles of Biological Circuits. See the book for more details and citations; see here for a review of most of the rest of the book.
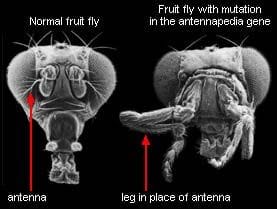
Fun fact: biological systems are highly modular, at multiple different scales. This can be quantified and verified statistically, e.g. by mapping out protein networks and algorithmically partitioning them into parts, then comparing the connectivity of the parts. It can also be seen more qualitatively in everyday biological work: proteins have subunits which retain their function when fused to other proteins, receptor circuits can be swapped out to make bacteria follow different chemical gradients, manipulating specific genes can turn a fly’s antennae into legs, organs perform specific functions, etc, etc.
On the other hand, systems designed by genetic algorithms (aka simulated evolution) are decidedly not modular. This can also be quantified and verified statistically. Qualitatively, examining the outputs of genetic algorithms confirms the statistics: they’re a mess.
So: what is the difference between real-world biological evolution vs typical genetic algorithms, which leads one to produce modular designs and the other to produce non-modular designs?
Kashtan & Alon tackle the problem by evolving logic circuits under various conditions. They confirm that simply optimizing the circuit to compute a particular function, with random inputs used for selection, results in highly non-modular circuits. However, they are able to obtain modular circuits using “modularly varying goals” (MVG).
The idea is to change the reward function every so often (the authors switch it out every 20 generations). Of course, if we just use completely random reward functions, then evolution doesn’t learn anything. Instead, we use “modularly varying” goal functions: we only swap one or two little pieces in the (modular) objective function. An example from the book:
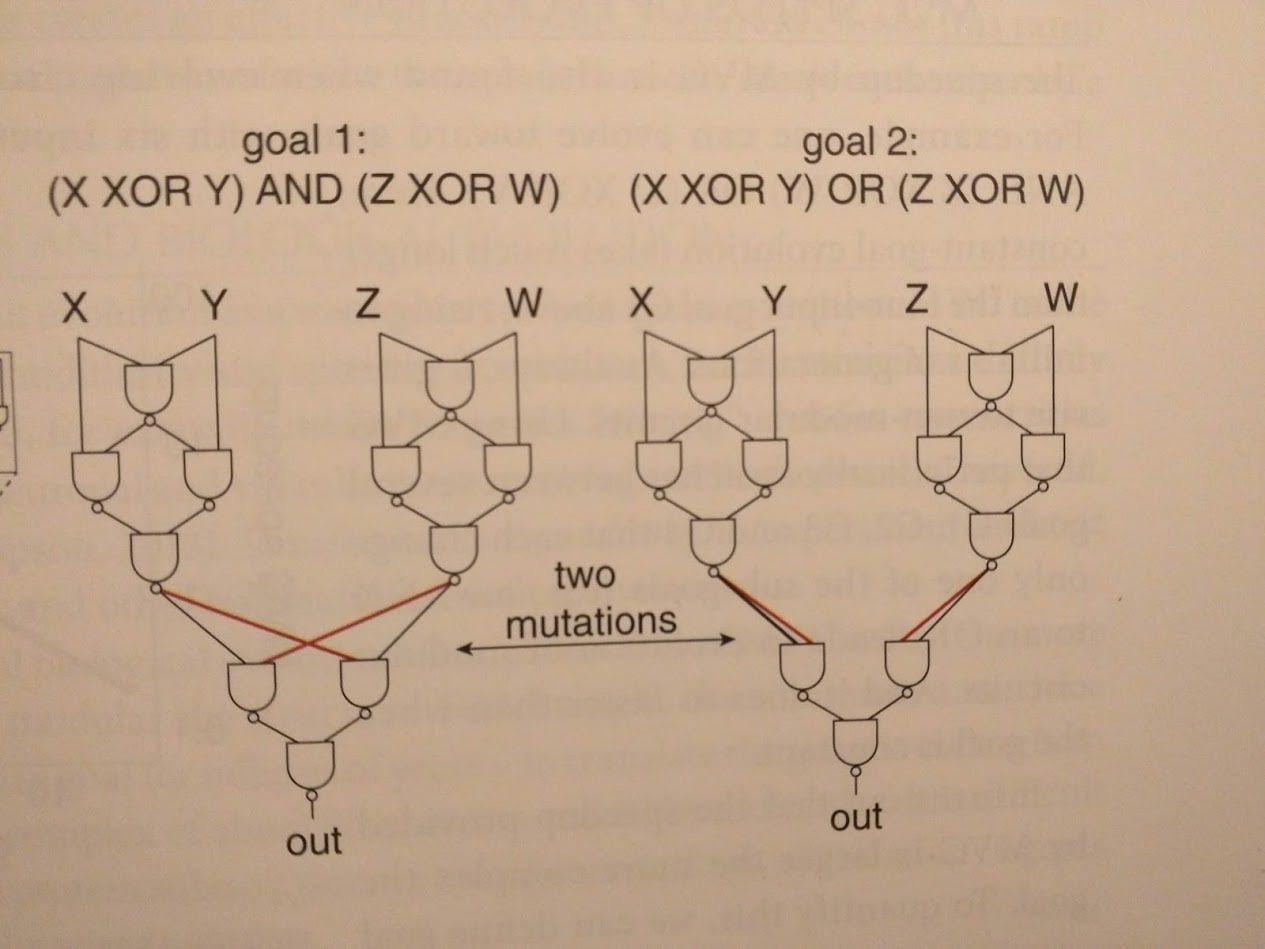
The upshot is that our different goal functions generally use similar sub-functions - suggesting that they share sub-goals for evolution to learn. Sure enough, circuits evolved using MVG have modular structure, reflecting the modular structure of the goals.
(Interestingly, MVG also dramatically accelerates evolution - circuits reach a given performance level much faster under MVG than under a fixed goal, despite needing to change behavior every 20 generations. See either the book or the paper for more on that.)
How realistic is MVG as a model for biological evolution? I haven’t seen quantitative evidence, but qualitative evidence is easy to spot. MVG as a theory of biological modularity predicts that highly variable subgoals will result in modular structure, whereas static subgoals will result in a non-modular mess. Alon’s book gives several examples:
- Chemotaxis: different bacteria need to pursue/avoid different chemicals, with different computational needs and different speed/energy trade-offs, in various combinations. The result is modularity: separate components for sensing, processing and motion.
- Animals need to breathe, eat, move, and reproduce. A new environment might have different food or require different motions, independent of respiration or reproduction - or vice versa. Since these requirements vary more-or-less independently in the environment, animals evolve modular systems to deal with them: digestive tract, lungs, etc.
- Ribosomes, as an anti-example: the functional requirements of a ribosome hardly vary at all, so they end up non-modular. They have pieces, but most pieces do not have an obvious distinct function.
To sum it up: modularity in the system evolves to match modularity in the environment.
The material here is one seed of a worldview which I've updated toward a lot more over the past year. Some other posts which involve the theme include Science in a High Dimensional World, What is Abstraction?, Alignment by Default, and the companion post to this one Book Review: Design Principles of Biological Circuits.
Two ideas unify all of these:
One major corollary of these two ideas is that goal-oriented systems will tend to evolve similar modular structures, reflecting the relevant parts of their environment. Systems to which this applies include organisms, machine learning algorithms, and the learning performed by the human brain. In particular, this suggests that biological systems and trained deep learning systems are likely to have modular, human-interpretable internal structure. (At least, interpretable by humans familiar with the environment in which the organism/ML system evolved.)
This post talks about some of the evidence behind this model: biological systems are indeed quite modular, and simulated evolution experiments find that circuits do indeed evolve modular structure reflecting the modular structure of environmental variations. The companion post reviews the rest of the book, which makes the case that the internals of biological systems are indeed quite interpretable.
On the deep learning side, researchers also find considerable modularity in trained neural nets, and direct examination of internal structures reveals plenty of human-recognizable features.
Going forward, this view is in need of a more formal and general model, ideally one which would let us empirically test key predictions - e.g. check the extent to which different systems learn similar features, or whether learned features in neural nets satisfy the expected abstraction conditions, as well as tell us how to look for environment-reflecting structures in evolved/trained systems.