Treat your obfuscated chains of thought like live bioweapons.
I've spoken to a few folks at NeurIPS that are training reasoning models against monitors for various reasons (usually to figure out how to avoid unmonitorable chain of thought). I had the impression not everyone was aware how dangerous these chain of though traces were:
- Make sure your obfuscated chains of thought are never used for LLM training!
If obfuscated reasoning gets into the training data, this could plausibly teach models how to obfuscate their reasoning. This seems potentially pretty bad (a bit like gain of function research). I'm not saying you shouldn't do the research, it's probably worth the risk. Just make sure to keep the rollouts away from training:
- Use e.g. the easy-dataset-share package (by TurnTrout et al.) to protect your dataset when you upload it somewhere (e.g. GitHub, HuggingFace).
- Don't use software that trains on your files when working with dangerous material (I think the free tiers of various AI products allow for training on user data).
As an example, consider the Claude 4 system card claiming that material from the Alignment Faking paper affected Claude's behaviour (discussion here).
Credit to pl...
glad people are noticing. it won't be enough to stop all leaks though, realistically.
it's fun how all the safety worries and intricate plans to prevent failure modes tend to get invalidated by "humans do the thing the bypasses the guardrails". e.g. for years people would say things like "of course we won't connect it to the internet/let it design novel viruses/proteins/make lethal autonomous weapons".
my guess is the law of less dignified failure has a lot of truth to it.
Here's an IMO under-appreciated lesson from the Geometry of Truth paper: Why logistic regression finds imperfect feature directions, yet produces better probes.
Consider this distribution of True and False activations from the paper:
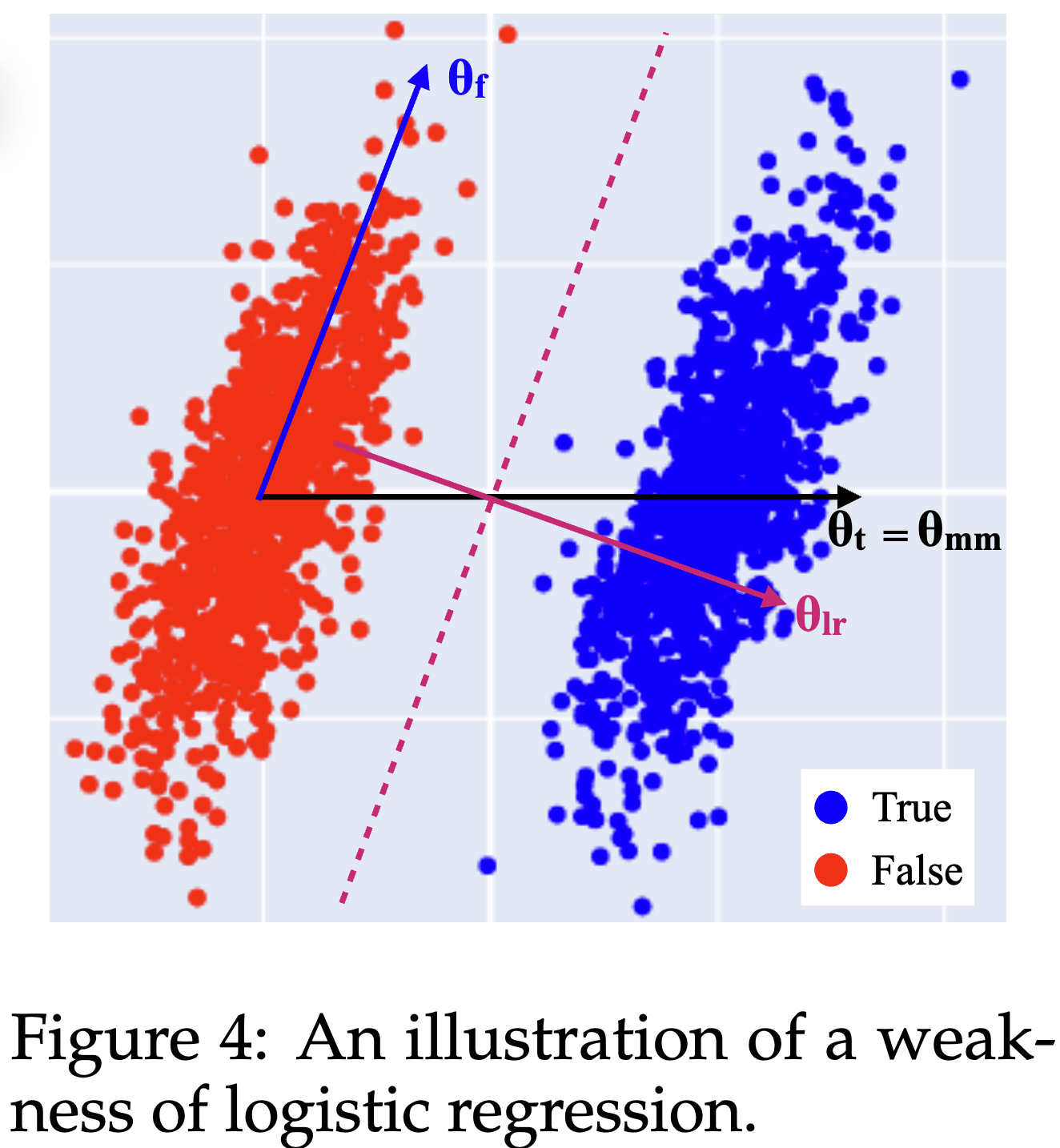
The True and False activations are just shifted by the Truth direction . However, there also is an uncorrelated but non-orthogonal direction along which the activations vary as well.
The best possible logistic regression (LR) probing direction is the direction orthogonal to the plane separating the two clusters, . Unintuitively, the best probing direction is not the pure Truth feature direction !
- This is a reason why steering and (LR) probing directions differ: For steering you'd want the actual Truth direction [1], while for (optimal) probing you want .
- It also means that you should not expect (LR) probing to give you feature directions such as the Truth feature direction.
The paper also introduces mass-mean probing: In the (uncorrelated) toy scenario, you can obtain the pure Truth feature direction from the difference between the distribution centroids .
- Contrastive methods (like mass
A fun side note, that probably isn't useful - I think if you shuffle the data across neurons (which effectively gets rid of the covariance amongst neurons), and then do linear regression, you will get theta_t.
This is a somewhat common technique in neuroscience analysis when studying correlation structure in neural reps and separability.
Edited to fix errors pointed out by @JoshEngels and @Adam Karvonen (mainly: different definition for explained variance, details here).
Summary: K-means explains 72 - 87% of the variance in the activations, comparable to vanilla SAEs but less than better SAEs. I think this (bug-fixed) result is neither evidence in favour of SAEs nor against; the Clustering & SAE numbers make a straight-ish line on a log plot.
Epistemic status: This is a weekend-experiment I ran a while ago and I figured I should write it up to share. I have taken decent care to check my code for silly mistakes and "shooting myself in the foot", but these results are not vetted to the standard of a top-level post / paper.
SAEs explain most of the variance in activations. Is this alone a sign that activations are structured in an SAE-friendly way, i.e. that activations are indeed a composition of sparse features like the superposition hypothesis suggests?
I'm asking myself this questions since I initially considered this as pretty solid evidence: SAEs do a pretty impressive job compressing 512 dimensions into ~100 latents, this ought to mean something, right?
But maybe all SAEs are doing is "dataset clustering" (the d...
I was having trouble reproducing your results on Pythia, and was only able to get 60% variance explained. I may have tracked it down: I think you may be computing FVU incorrectly.
https://gist.github.com/Stefan-Heimersheim/ff1d3b92add92a29602b411b9cd76cec#file-clustering_pythia-py-L309
I think FVU is correctly computed by subtracting the mean from each dimension when computing the denominator. See the SAEBench impl here:
https://github.com/adamkarvonen/SAEBench/blob/5204b4822c66a838d9c9221640308e7c23eda00a/sae_bench/evals/core/main.py#L566
When I used your FVU implementation, I got 72% variance explained; this is still less than you, but much closer, so I think this might be causing the improvement over the SAEBench numbers.
In general I think SAEs with low k should be at least as good as k means clustering, and if it's not I'm a little bit suspicious (when I tried this first on GPT-2 it seemed that a TopK SAE trained with k = 4 did about as well as k means clustering with the nonlinear argmax encoder).
Here's my clustering code: https://github.com/JoshEngels/CheckClustering/blob/main/clustering.py
this seems concerning.
I feel like my post appears overly dramatic; I'm not very surprised and don't consider this the strongest evidence against SAEs. It's an experiment I ran a while ago and it hasn't changed my (somewhat SAE-sceptic) stance much.
But this is me having seen a bunch of other weird SAE behaviours (pre-activation distributions are not the way you'd expect from the superposition hypothesis h/t @jake_mendel, if you feed SAE-reconstructed activations back into the encoder the SAE goes nuts, stuff mentioned in recent Apollo papers, ...).
Reasons this could be less concerning that it looks
- Activation reconstruction isn't that important: Clustering is a strong optimiser -- if you fill a space with 16k clusters maybe 90% reconstruction isn't that surprising. I should really run a random Gaussian data baseline for this.
- End-to-end loss is more important, and maybe SAEs perform much better when you consider end-to-end reconstruction loss.
- This isn't the only evidence in favour of SAEs, they also kinda work for steering/probing (though pretty badly).
What's up with different LLMs generating near-identical answers?
TL,DR: When you ask the same question you often get the same answer. These feel way more deterministic that I'd expect based on webtext probabilities (pre-training). Of course post-training hugely influences this kind of thing (mode collapse?), it's just more striking that I had thought.
Following this post, I asked "Tell me a funny joke":
- GPT o3: "Why don’t scientists trust atoms anymore? Because they make up everything! 😁 Want to hear another one, or maybe something in a different style?"
- Claude 3.7 Sonnet (thinking): "Why don't scientists trust atoms? Because they make up everything!"
- Gemini 2.5 Pro (preview): "Why don't scientists trust atoms? Because they make up everything!"
What's going on here? Asking for five more jokes from each model, I only get one overlapping one ("Why don't skeletons fight each other? They don't have the guts.", from Claude and Gemini).
I've also tried "Tell me your #1 fact about the ocean":
- GPT o3: "More than half of every breath you take is thanks to the ocean — tiny, photosynthetic plankton drifting in sun-lit surface waters generate at least 50 % of Earth’s oxygen supply."
- Claude: "My
Why can't the mode-collapse just be from convergent evolution in terms of what the lowest-common denominator rater will find funny? If there are only a few top candidates, then you'd expect a lot of overlap. And then there's the very incestuous nature of LLM training these days: everyone is distilling and using LLM judges and publishing the same datasets to Hugging Face and training on them. That's why you'll ask Grok or Llama or DeepSeek-R1 a question and hear "As an AI model trained by OpenAI...".
Collection of some mech interp knowledge about transformers:
Writing up folk wisdom & recent results, mostly for mentees and as a link to send to people. Aimed at people who are already a bit familiar with mech interp. I've just quickly written down what came to my head, and may have missed or misrepresented some things. In particular, the last point is very brief and deserves a much more expanded comment at some point. The opinions expressed here are my own and do not necessarily reflect the views of Apollo Research.
Transformers take in a sequence of tokens, and return logprob predictions for the next token. We think it works like this:
- Activations represent a sum of feature directions, each direction representing to some semantic concept. The magnitude of directions corresponds to the strength or importance of the concept.
- These features may be 1-dimensional, but maybe multi-dimensional features make sense too. We can either allow for multi-dimensional features (e.g. circle of days of the week), acknowledge that the relative directions of feature embeddings matter (e.g. considering days of the week individual features but span a circle), or both. See also Jake Mendel's
Against multi-page forms.
I dislike questionnaires / forms split into multiple pages where I can't see the full length of the questionnaire without starting to fill it in. I usually want to know how much effort a survey takes before deciding to invest time into filling it out, or to plan how much time to allocate.
Example: Martian's interpretability grants form (6 pages, edit: but they said they’ll fix it). I cant't see how much effort an application is, so I might not fill in the first 3 pages because I worry that the last 3 pages will be too much effort to be worth the time.
Alternative: Open Philanthropy's RFP EOI form (now closed) was a single-page form. I could see how much total effort was required to apply, and decide whether it was worth the expected value.
Edit: Obviously, if you're running an experiment / interview / test where it's important the subject doesn't see the next page before filling out the first page, this is fine.
PSA: People use different definitions of "explained variance" / "fraction of variance unexplained" (FVU)
is the formula I think is sensible; the bottom is simply the variance of the data, and the top is the variance of the residuals. The indicates the norm over the dimension of the vector . I believe it matches Wikipedia's definition of FVU and R squared.
is the formula used by SAELens and SAEBench. It seems less principled, @Lucius Bushnaq and I couldn't think of a nice quantity it corresponds to. I think of it as giving more weight to samples that are close to the mean, kind-of averaging relative reduction in difference rather than absolute.
A third version (h/t @JoshEngels) which computes the FVU for each dimension independently and then averages, but that version is not used in the context we're discussing here.
In my recent comment I had computed my own , and compared it to FVUs from SAEBench (which used ) and obtained nonsense results.
Curiously the two definitions seem to be approximately proportional—below I show the pe...
Memorization in LLMs is probably Computation in Superposition (CiS, Vaintrob et al., 2024).
CiS is often considered a predominantly theoretical concept. I want to highlight that most memorization in LLMs is probably CiS. Specifically, the typical CiS task of "compute more AND gates than you have ReLU neurons" is exactly what you need to memorize lots of facts. I'm certainly not the first one to say this, but it also doesn't seem common knowledge. I'd appreciate pushback or references in the comments!
Consider the token “Michael”. GPT-2 knows many things about Michael, including a lot of facts about Michael Jordan and Michael Phelps, all of which are relevant in different contexts. The model cannot represent all these in the embedding of the token Michael (conventional superposition, Elhagge et al., 2022); in fact—if SAEs are any indication—the model can only represent about 30-100 features at a time.
So this knowledge must be retrieved dynamically. In the sentence “Michael Jordan plays the sport of”, a model will consider the intersection of Michael AND Jordan AND sport, resulting in basketball. Folk wisdom is that this kind of memorization is implemented by the MLP blocks in a Transf...
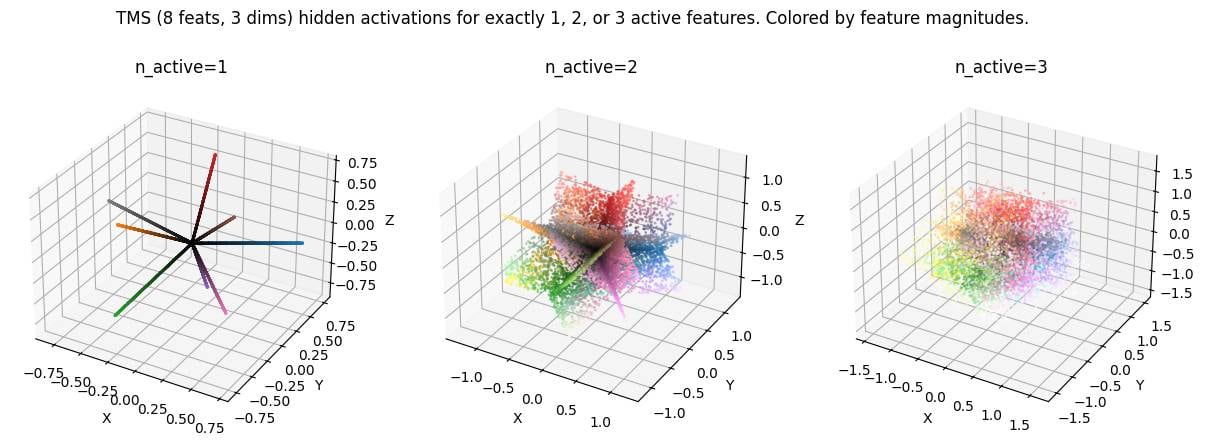
LLM activation space is spiky. This is not a novel idea but something I believe many mechanistic interpretability researchers are not aware of. Credit to Dmitry Vaintrob for making this idea clear to me, and to Dmitrii Krasheninnikov for inspiring this plot by showing me a similar plot in a setup with categorical features.
Under the superposition hypothesis, activations are linear combinations of a small number of features. This means there are discrete subspaces in activation space that are "allowed" (can be written as the sum of a small number of features), while the remaining space is "disallowed" (require much more than the typical number of features).[1]
Here's a toy model (following TMS, total features in -dimensional activation space, with features allowed to be active simultaneously). Activation space is made up of discrete -dimensional (intersecting) subspaces. My favourite image is the middle one () showing planes in 3d activation space because we expect in realistic settings.
( in the plot corresponds to here. Code here.)
This picture predicts that interpolating between two activations should take you out-of-distribution ...
Is weight linearity real?
A core assumption of linear parameter decomposition methods (APD, SPD) is weight linearity. The methods attempt to decompose a neural network parameter vector into a sum of components such that each component is sufficient to execute the mechanism it implements.[1] That this is possible is a crucial and unusual assumption. As counter-intuition consider Transcoders, they decompose a 768x3072 matrix into 24576 768x1 components which would sum to a much larger matrix than the original.[2]
Trivial example where weight linearity does not hold: Consider the matrix in a network that uses superposition to represent 3 features in two dimensions. A sensible decomposition could be to represent the matrix as the sum of 3 rank-one components
If we do this though, we see that the components sum to more than the original matrix
The decomposition doesn’t work, and I can’t find any other decomposition that makes sense. However, APD claims that this matrix should be described as a sin...
I don't like the extensive theming of the frontpage around If Anyone Builds It, Everyone Dies.
The artwork is distracting. I just went on LW to create a new draft, got distracted, clicked on the website, and spent 3 minutes reporting a bug. I expect this is intended to some degree, but it feels a little "out to get you" to me.
Edit: The mobile site looks quite bad too (it just looks like unintended dark mode)
Why I'm not too worried about architecture-dependent mech interp methods:
I've heard people argue that we should develop mechanistic interpretability methods that can be applied to any architecture. While this is certainly a nice-to-have, and maybe a sign that a method is principled, I don't think this criterion itself is important.
I think that the biggest hurdle for interpretability is to understand any AI that produces advanced language (>=GPT2 level). We don't know how to write a non-ML program that speaks English, let alone reason, and we have no idea how GPT2 does it. I expect that doing this the first time is going to be significantly harder, than doing this the 2nd time. Kind of how "understand an Alien mind" is much harder than "understand the 2nd Alien mind".
Edit: Understanding an image model (say Inception V1 CNN) does feel like a significant step down, in the sense that these models feel significantly less "smart" and capable than LLMs.
List of some larger mech interp project ideas (see also: short and medium-sized ideas). Feel encouraged to leave thoughts in the replies below!
Edit: My mentoring doc has more-detailed write-ups of some projects. Let me know if you're interested!
What is going on with activation plateaus: Transformer activations space seems to be made up of discrete regions, each corresponding to a certain output distribution. Most activations within a region lead to the same output, and the output changes sharply when you move from one region to another. The boundaries seem...
Are the features learned by the model the same as the features learned by SAEs?
TL;DR: I want true features model-features to be a property of the model weights, and to be recognizable without access to the full dataset. Toy models have that property. My “poor man’s model-features” have it. I want to know whether SAE-features have this property too, or if SAE-features do not match the true features model-features.
Introduction: Neural networks likely encode features in superposition. That is, features are represented as directions in...
List of some medium-sized mech interp project ideas (see also: shorter and longer ideas). Feel encouraged to leave thoughts in the replies below!
Edit: My mentoring doc has more-detailed write-ups of some projects. Let me know if you're interested!
Toy model of Computation in Superposition: The toy model of computation in superposition (CIS; Circuits-in-Sup, Comp-in-Sup post / paper) describes a way in which NNs could perform computation in superposition, rather than just storing information in superposition (TMS). It would be good to have some actually trai...
I've heard people say we should deprioritise fundamental & mechanistic interpretability[1] in short-timelines (automated AI R&D) worlds. This seems not obvious to me.
The usual argument is
- Fundamental interpretability will take many years or decades until we "solve interpretability" and the research bears fruits.
- Timelines are short, we don't have many years or even decades.
- Thus we won't solve interpretability in time.
But this forgets that automated AI R&D means we'll have decades of subjective research-time in months or years of wall-clock t...
Edit: I feel less strongly following the clarification below. habryka clarified that (a) they reverted a more disruptive version (pixel art deployed across the site) and (b) that ensuring minimal disruption on deep-links is a priority.
I'm not a fan of April Fools' events on LessWrong since it turned into the de-facto AI safety publication platform.
We want people to post serious research on the site, and many research results are solely hosted on LessWrong. For instance, this mech interp review has 22 references pointing to lesswrong.com (along with 22 furt...
It's always been a core part of LessWrong April Fool's that we never substantially disrupt or change the deep-linking experience.
So while it looks like a lot of going on today, if you get linked directly to an article, you will basically notice nothing different. All you will see today are two tiny pixel-art icons in the header, nothing else. There are a few slightly noisy icons in the comment sections, but I don't think people would mind that much.
This has been a core tenet of all April Fool's in the past. The frontpage is fair game, and April Fool's jokes are common for large web platforms, but it should never get in the way of accessing historical information or parsing what the site is about, if you get directly linked to an author's piece of writing.
List of some short mech interp project ideas (see also: medium-sized and longer ideas). Feel encouraged to leave thoughts in the replies below!
Edit: My mentoring doc has more-detailed write-ups of some projects. Let me know if you're interested!
Directly testing the linear representation hypothesis by making up a couple of prompts which contain a few concepts to various degrees and test
- Does the model indeed represent intensity as magnitude? Or are there separate features for separately intense versions of a concept? Finding the right prompts is tricky, e.g.
We want two different kinds of probes in a white-box monitoring setup.
Say we want to monitor LLM queries for misuse or various forms of misalignment. We'll probably use a hierarchical monitoring setup (e.g. Hua et al.) with a first cheap stage, and a more expensive second stage.
When I talk to people about model internals-based probes, they typically think of probes as the first-stage filter only.
Instead, we should use probes anywhere where they're helpful. I expect we want:
- A first-stage probe (applied to all samples), optimised for low cost & high reca
Why I'm not that hopeful about mech interp on TinyStories models:
Some of the TinyStories models are open source, and manage to output sensible language while being tiny (say 64dim embedding, 8 layers). Maybe it'd be great to try and thoroughly understand one of those?
I am worried that those models simply implement a bunch of bigrams and trigrams, and that all their performance can be explained by boring statistics & heuristics. Thus we would not learn much from fully understanding such a model. Evidence for this is that the 1-layer variant, which due t...
Has anyone tested whether feature splitting can be explained by composite (non-atomic) features?
- Feature splitting is the observation that SAEs with larger dictionary size find features that are geometrically (cosine similarity) and semantically (activating dataset examples) similar. In particular, a larger SAE might find multiple features that are all similar to each other, and to a single feature found in a smaller SAE.
- Anthropic gives the example of the feature " 'the' in mathematical prose" which splits into features " 'the' in mathematics
I think we should think more about computation in superposition. What does the model do with features? How do we go from “there are features” to “the model outputs sensible things”? How do MLPs retrieve knowledge (as commonly believed) in a way compatible with superposition (knowing more facts than number of neurons)?
This post (and paper) by @Kaarel, @jake_mendel, @Dmitry Vaintrob (and @LawrenceC) is the kind of thing I'm looking for, trying to lay out a model of how computation in superposition could work. It makes somewhat-concrete predictions ...