Suppose you want to decrease your risk of heart disease. The conventional advice goes something like this:
- Eat a healthier diet with less LDL-cholesterol raising foods
- Exercise more
- Keep your blood sugar under control
- Don’t smoke, don’t sit too much and don't take 400mg of methamphetamine on a regular basis
An alternative strategy might be some kind of genetic intervention. For example, an active clinical trial by Verve Therapeutics aims to treat individuals with inherited high cholesterol by editing the PCSK9 gene.
These trials almost always start the same: there’s some rare disorder caused by a single gene. We have a strong mechanical understanding of how the gene causes the disorder. We use an animal model with an analogous disorder and show that by changing the gene we fix or at least ameliorate the condition.
This is the traditional approach. And despite being slow and limited in scope, it occasionally produces results like Casgevy, a CRISPR-based treatment for sickle cell and beta thallasemia which was approved by the UK in mid-November.
It might cost several million dollars. But it cures sickle cell! That has to count for something.
Most diseases, however, are not like sickle cell or beta thalassemia. They are not caused by one gene. They are caused by the cumulative effects of thousands of genes plus environmental factors like diet and lifestyle.
If we actually want to treat these disorders, we need to start thinking about biology (and genetic treatments) differently.
Black Box Biology
I think the conventional approach to genes and disorders is fundamentally stupid. In seeking absolute certainty about cause and effect, it limits itself to a tiny niche with limited importance. It’s as if machine learning researchers decided that the best way to build a neural network was to hand tune model parameters based on their intricate knowledge of feature representations.
You don’t need to understand the mechanism of action. You don’t need an animal model of disease. You just need a reasonable expectation that changing a genetic variant will have a positive impact on the thing you care about.
And guess what? We already have all that information.
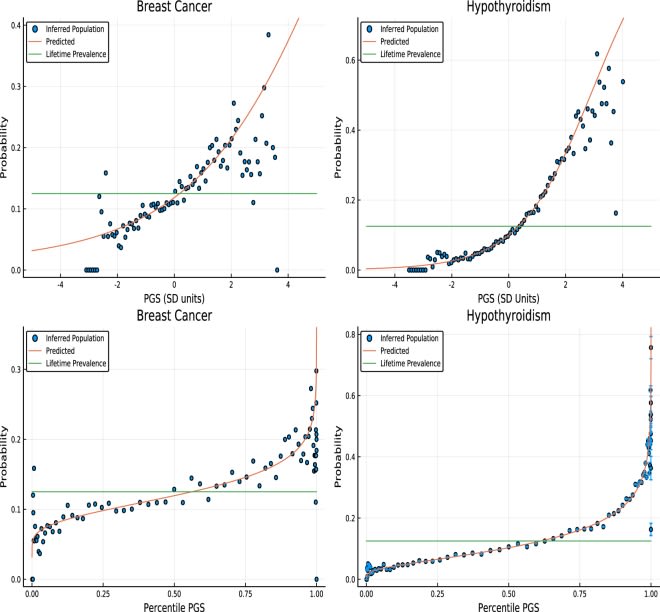
We’ve been conducting genome-wide association studies for over a decade. A medium-sized research team can collect data from 180,000 diabetics and show you 237 different spots in the genome that affect diabetes risk with a certainty level of P < 5*10^-9!
In expectation, editing all those variants could decrease someone’s diabetes risk to negligible levels.
I predict that in the next decade we are going to see a fundamental shift in the way scientists think about the relationship between genes and traits. The way treatments change outcomes is going to become a black box and everyone will be fine with it because it will actually work.
We don’t need to understand the mechanism of action. We don’t need to understand the cellular pathway. We just need enough data to know that when we change this particular base pair from an A to a G, it will reduce diabetes risk by 0.3%.
That’s enough.
Thanks, this is exciting and inspiring stuff to learn about!
I guess another thing I'm wondering about, is how we could tell apart genes that impact a trait via their ongoing metabolic activities (maybe metabolic is not the right term... what I mean is that the gene is being expressed, creating proteins, etc, on an ongoing basis), versus genes that impact a trait via being important for early embryonic / childhood development, but which aren't very relevant in adulthood. Genes related to intelligence, for instance, seem like they might show up with positive scores in a GWAS, but their function is confined to helping unfold the proper neuron connection structures during fetal development, and then they turn off, so editing them now won't do anything. Versus other genes that affect, say, what kinds of cholesterol the body produces, seem more likely to have direct impact via their day-to-day operation (which could be changed using a CRISPR-like tool).
Do we have any way of distinguishing the one type of genes from the other? (Maybe we can just look at living tissue and examine what genes are expressed vs turned off? This sounds hard to do for the entire genome...) Or perhaps we have reason to believe something like "only 20% of genes are related to early development, 80% handle ongoing metabolism, so the GWAS --> gene therapy pipeline won't be affected too badly by the dilution of editing useless early-development genes"?